BioBlitz Taxonomy Enhancements
Additional support for users of an existing database system
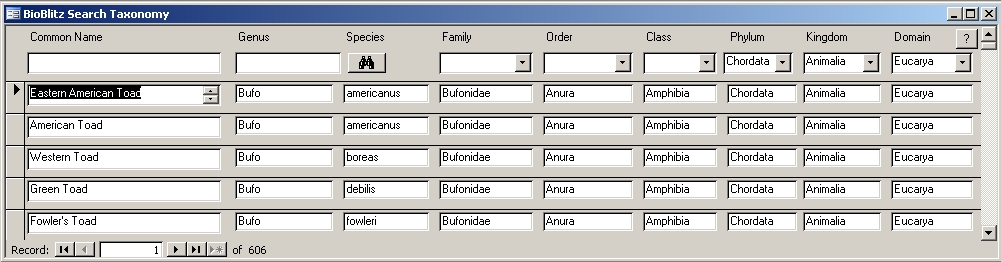
Screen capture of database providing taxonomic enhancements for an existing BioBlitz database. Click on image to enlarge.
This screen demonstrating selection for vertebrates (Phylum Chordata) while narrowing in on amphibians to find the genus and species for American Toad (even recognizing multiple common names).
Why was this developed?
In 2007, Jug Bay Wetlands Sanctuary planned a BioBlitz, a 24-hour event to find and identify the variety of species at their site. Instead of developing a new database system to manage and analyze the results from the BioBlitz, we looked for existing solutions. At that time, the All Taxa Biodiversity (ATBI) database was available as a "desktop version" in Microsoft Access. Information about ATBI is provided on the Discover Life in America website, but it appears the Access database is no longer available.
The Jug Bay BioBlitz included heavy involvement with citizen scientists. The ATBI seems to have been designed for use by personnel with more taxonomic expertise and a more formal research structure. However, the database design was very good and it more than met the Jug Bay requirements. We did several things to make it easier for the volunteers.
How does it work?
Design strategy
Since ATBI required entry of data based on the genus and species, Jeff developed an interface to a new database to allow searching at a level higher than genus. A new database was created instead of modifying the ATBI database to maintain the integrity of the ATBI database and to avoid the need to license development tools.
The basis for the taxonomic data is the Integrated Taxonomic Information System (ITIS). That data was downloaded and put into tables in the new, supplemental database.
User Interface
The main screen was designed to allow searching to find the genus and species based on any part of the common name or whatever taxonomic information was known. For example, the person could search for "amphibians" by selecting "amphibia" in the Class drop down list. They could "pickerel" in the common name and the results would show several species of pickerel fish as well as pickerel frog.
The list boxes initially list all names at that level, for example "class" displays classes for all plants, animals, fungi, and microbes. However, when a higher level is chosen, for example the animal kingdom (Animalia) then the lower level list boxes would only be the ones for animals. The user can also double-click on the list box to clear the selection at that level (and lower levels)
The list can be sorted alphabetically (A to Z or Z to A) by clicking on the label at the top of the column ("Common Name," "Genus," etc.).
Once the user found the genus and species, these could be copied and pasted into the ATBI data entry screen.
Technical details
The ability to "link" tables between Access databases made data integration with the ATBI database easy. In particular, when searching for a species, the user could search the entire ITIS species list or just the species already added to the ATBI database. Since there was significant overlap of species that were pre-loaded in the Great Smoky database with the Jug Bay site, the majority of volunteer searches would be for species already in the ATBI database. Species from spreadsheets listing species already known to be found at Jug Bay were also added to the ATBI database before the BioBlitz.
Initial results
The volunteers entering the data greatly appreciated the easier method for looking up scientific names. The combination of ATBI with the taxonomic supplement was used for several BioBlitzes at three locations for several years. The 2007 results and 2009 results are available on the Jug Bay web site.
Conclusions
- Using the ATBI database provided a good underlying database without having to do significant development and was available within the short lead time for the event.
- Some groups of plants/animals were not identified to the species level. For example, stream macroinvertebrates were only identified to the class or family. This could be handled by adding "unknown" family, genus, and species. However, simply creating a single "unknown" at each taxonomic level would not work with the existing ATBI reporting and analysis. A spreadsheet was used to handle the data that was not easily added to the database.
- While not a surprise to biologists, but not obvious to computer scientists, the taxonomic structure/names change over time. The ITIS database uses good design principles and assigns a unique identifier (number) to each species. This means that the name(s) can change without "breaking" the existing data. However, this could require on-going maintenance of the taxonomic database included within a BioBlitz system. Since the supplemental taxonomic database described above simply imported data from ITIS, this maintenance would be fairly easy.
- Technology has changed a lot since this was created in 2007. With increased Internet access and better search tools, it is not clear that the supplemental database now provides as much benefit in comparison with current alternatives as it did in 2007. For example, the ITIS web site search page now implements richer search capabilities that could substitute for this custom design from 2007.